GeneratePeptides
In order to use ProteinFishing, the FishXDB has to be installed and prepared as explained in our tutorial here. Moreover the FishXDB has to be filled with fragments obtained by the digestion of PDB structures you want to fish up.
To do so you have to use the GeneratePeptides subroutine as follow with each one of the structures you want to digest:
ProteinFishing
The basic run (using default parameters) of ProteinFishing looks like this:
>modelX --command=ProteinFishing --pdb=yourPdb.pdb --residue-begin=1 --residue-end=6 --hook-mol=C --fisher-mol=A --database-name=FishXDB --database-server=localhost --database-user=modelx --database-pass=modelx
To play around with additional ProteinFishing parameters please refer to the command section here.
Your fishes
The output of a ProteinFishing run is contained in a directory named with the word "fishnet" and the name of your input PDB structure. In this folder you will find another directory named with the name of the Hook molecule, the starting and ending positions of the Hook window you are using. Within this folder you can find your ProteinFishing models and a Summary file of your ProteinFishing run. Models are named with yourPdb.pdb name, the name of the PDB model that have been fished and a counter starting from zero.
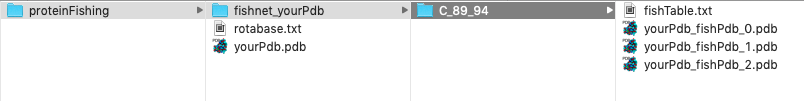
ProteinFishing summary file
The proteinFishing summary file is always named as "fishTable.txt", contains informations on your ProteinFishing run and looks like this:
yourPdb_fishPdb_0 19 2 -26
yourPdb_fishPdb_1 15 1 -17.7
yourPdb_fishPdb_2 16 1 -98.9
FishingLure
The FishingLure command allows you to run ProteinFishing without determining a Hook window over the Hook molecule, automatically determine multiple windows and launch an individual ProteinFishing process for each one. The basic run of FishingLure looks like this:
>modelX command=FishingLure --pdb=yourPdb.pdb --fisher-mol=A --hooks-length=6 --max-threads=1 --database-name=FishXDB --database-server=localhost --database-user=modelx --database-pass=modelx
To play around with additional FishingLure parameters please refer to the command section here.

